 Scripps XtalView WWW Page
Scripps XtalView WWW Page
last updated Aug 18, 2000
Mirror sites for these pages - use these to speed up download times:
Across the Atlantic: CCP14
site in Daresbury, UK
Down under: CCP14
mirror in Melbourne, Australia
NEW!!
XtalView for Mac OS X README Download
Hardware
stereo for LINUX, DEC alpha's
Xfit 4 now works with
probe
The second edition of Practical Protein Crystallography is available
Want to convert a CNS map to XtalView? Check out Christopher Putnam's
CNS2FSFOUR
 XtalView
3.2
XtalView
3.2
XtalView is a crystallographic software package for fitting electron density
maps and solving structures by MIR and MAD
-
Authors: Duncan
E. McRee, Alex Shah, Mark Israel, and Michael Pique
-
Distribution
XtalView 3.2 - New features
-
Xfit
-
Support for ultra high resolution refinement
-
Anisotropic thermal parameters - reads writes U's and displays thermal
probabilities
-
Improved disorder handling - can edit each side independently or
together
-
SHELX-97 CIF Structure
factor files (LIST 6)
-
B-splines - improves accuracy of interpolation without increasing
memory use
-
Arbitrary contour grids
-
Improved geometry refinement
-
One-step refinements on Autofit menu:
-
Refine Region - click a residue and then choose this to refine a
residue plus its two neighbours, hit spacebar until finished
-
Refine Range - click two endpoints, issue command, hit spacebar
until finished
-
Refine-while-fitting - lets you drag
atoms while the geometry and fit to density is being continuously updated
-
Phi-psi optimized to Ramachandran plot
-
Chi1 angles optimized by residue type
-
SigmaA maps - SigmaA 2mFo-Dfc and Fo-mFc maps are computed on the
fly - combined with the "shake" option in SfCalc window this can greatly
reduce phase bias
-
Raster3D interface
- Xfit can call Raster3D directly for high quality rendering of map and
model
-
Three dictionaries available with a click of the button on the Model
window
-
No hydrogens
-
Polar hydrogens
-
Full hydrogens
-
More conformers added to dictionaries
-
Real-time contour levels - contours are adjusted interactively as
you change levels
-
Model geometry error list - finds geometry errors - click an error
to center it
-
Model window
-
Copy into buffer as well as cut
-
Renumber chain
-
Reverse chain
-
Sort by sequence numbers
-
Make C-terminus (adds second oxygen)
-
Ribosyme - insert now makes peptide bond to previous/next residue
automatically
-
Atom level editing - remove an atom - change B/occupancy
-
OS
-
IRIX 6
-
LINUX "unwanted features" removed
These features are still there
-
Xfit
-
Fits Proteins, Nucleic acids, carbohydrates and any arbitrary organic
molecule
-
On the fly omit maps
-
Built in FFT - use phases instead of maps
-
Built in Structure Factor Calculations - update density to reflect
model changes
-
Automated
tracing
-
20 levels of undo for model changes
-
Cut and paste between models
-
Split residue around active torsion
-
All major UNIX OS's supported (including LINUX)!
-
Refines against map and geometry in real-time
 Raster3D
Support added in XtalView 3.2
Raster3D
Support added in XtalView 3.2
The new version of XtalView writes Raster3D input. You will need XtalView
3.2+ and Raster3D 2.3+
.
To
order XtalView click here
These are late releases version of xfit for those who can't wait. Download
the appropiate one and and put in the same directory as your current xfit
(can be found by issuing a "which xfit" command). Note: these are unsupported
and won't run correctly without the rest of XtalView. Other machine types,
contact us.
Hint: right-click the URL and pull down to Save Link As...
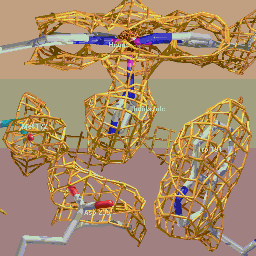
The title image was made by contouring density in xfit and rendering
with Molecular Images
software.
We have had 1066 visits since we started counting 8/1/97 (780 from
8/1-12/17/97)
Contact: [email protected] for XtalView help and [email protected]
(Duncan E. McRee) for comments on these pages.
This page best when viewed with a web browser
 XtalView
3.2
XtalView
3.2
 Scripps XtalView WWW Page
Scripps XtalView WWW Page
 XtalView
3.2
XtalView
3.2
 Raster3D
Support added in XtalView 3.2
Raster3D
Support added in XtalView 3.2