Now read the E-mails on the subject via the Rietveld Users' Mailing List to get a good feel for
what is going and some issues you can expect when doing this type of refinement
Date: Mon, 02 Dec 2002 17:29:27 +0000
To: [email protected]
From: Lachlan Cranswick [[email protected]]
Subject: RIET: GSAS query on importing Protein PDB atom positions?
For importing a protein PDB file into GSAS via EXPEDT. How is this
done?
There is a menu option for importing BNL PDB format in EXPEDT
via K L A I B - but there seems to be no prompt for the
filename - and the following error is given if you try using
the I B menu option?
Give atom editing command (,$,I,S,X) >i
Command structure for inserting an atom
I s - enter atom with sequence number "s"
I N - enter atom with next sequence number
I B - Read atoms from BNL PDB format
I R - Read atoms from non-GSAS file or from another EXP file
Phase No. 1; Phase has 0 atoms; Title: blah
Give atom editing command (,$,I,S,X) >i B
PDB format only allowed for protein phase type
*** No atoms found for B - ERROR - ***
Phase No. 1; Phase has 0 atoms; Title: blah
Give atom editing command (,$,I,S,X) >
==================
If you type I R - a GSAS EXP file is prompted for.
Thanks in advance,
Lachlan.
-----------------------
Lachlan M. D. Cranswick
Collaborative Computational Project No 14 (CCP14)
for Single Crystal and Powder Diffraction
Birkbeck University of London and Daresbury Synchrotron Laboratory
Postal Address: CCP14 - School of Crystallography,
Birkbeck College,
Malet Street, Bloomsbury,
WC1E 7HX, London, UK
Tel: (+44) 020 7631 6850 Fax: (+44) 020 7631 6803
E-mail: [email protected] Room: B091
WWW: http://www.ccp14.ac.uk/
Date: Mon, 02 Dec 2002 18:55:26 +0100
To: [email protected]
From: Jonathan WRIGHT [[email protected]]
Subject: Re: RIET: GSAS query on importing Protein PDB atom positions?
Lachlan,
Did you toggle the phase flags to say this is macromolecule (p p m 1 d) ?
The hint was in you mail ;-)
>PDB format only allowed for protein phase type
Although I thought I read somewhere that recent versions had this
restriction removed... wishful thinking perhaps?
Cheers,
Jon
From: "Bob Von Dreele" [[email protected]]
Subject: Re: RIET: GSAS query on importing Protein PDB atom positions?
To: [email protected]
Date: Mon, 02 Dec 2002 10:44:40 -0800
Hi Lachlan,
The phase type must be "macromolecular" for the "I B"
option to work. Sometime I'll fix this so
nonmacromolecular structures can be read too.
Bob
Date: Mon, 02 Dec 2002 18:12:26 +0000
To: [email protected]
From: Lachlan Cranswick
Subject: Re: RIET: GSAS query on importing Protein PDB atom positions?
Thanks Bob and Jon for the info (I set this file up with
EXPGUI as part of the Le Bail fitting - so did not
appreciate the "phase type"):
As per previous Emails, change phase type:
Y P P
M 1 !modify phase type for phase 1
D !macromolecular structure
Insert PDB File:
Y L A
I B
file.pdb
(and answer the questions)
Would you normally also import the water/HOH molecules, disordered
atoms and hydrogens?
(though the PDB file I seem to have does not include the hydrogens)
----
Next query if there is the tolerance for it - how do you enter
the "solvent scattering"? And are there any other settings
that are important - that could be missed by someone more
used to refining inorganics?
I take it after the structure in happilly imported, it is
just a matter of running all the restraints macros in
the c:\gsas\macros directory (except C60)
Cheers,
Lachlan.
-----------------------
Lachlan M. D. Cranswick
Collaborative Computational Project No 14 (CCP14)
for Single Crystal and Powder Diffraction
Birkbeck University of London and Daresbury Synchrotron Laboratory
Postal Address: CCP14 - School of Crystallography,
Birkbeck College,
Malet Street, Bloomsbury,
WC1E 7HX, London, UK
Tel: (+44) 020 7631 6850 Fax: (+44) 020 7631 6803
E-mail: [email protected] Room: B091
WWW: http://www.ccp14.ac.uk/
From: "Bob Von Dreele" [[email protected]]
Subject: Re: RIET: GSAS query on importing Protein PDB atom positions?
To: [email protected]
Date: Mon, 02 Dec 2002 12:26:13 -0800
Hi Lachlan (& everyone else!),
See below...
Bob
On Mon, 02 Dec 2002 18:12:26 +0000
Lachlan Cranswick wrote:
>
>Thanks Bob and Jon for the info (I set this file up with
>EXPGUI as part of the Le Bail fitting - so did not
>appreciate the "phase type"):
>
>As per previous Emails, change phase type:
>
>Y P P
>M 1 !modify phase type for phase 1
>D !macromolecular structure
>
>Insert PDB File:
>
>Y L A
>I B
>file.pdb
>(and answer the questions)
One important point is that in a multiphase mixture the
protein must be phase #1. That is the only one that can be
"macromolecular". Current limits are 500 atoms for
"ordinary" phases and 5000 atoms for macromolecular ones.
>Would you normally also import the water/HOH molecules,
>disordered
>atoms and hydrogens?
>(though the PDB file I seem to have does not include the
>hydrogens)
"Normally" not for powder data. Single crystal protein
folks seem only to add waters with higher resolution data
(dmin ~ 2.5A or better). Some PDB files do have H-atoms. I
change the UISO's for all the atoms to 0.3 which is about
average for protein atoms. Powder data isn't good enough
to do UISO's in a refinement even when all constrained to
be identical.
>Next query if there is the tolerance for it - how do you
>enter
>the "solvent scattering"?
Solvent scattering is set in the form factor menu under
"S". Start with A=5 & U=1. These can be refined.
>And are there any other
>settings
>that are important - that could be missed by someone more
>used to refining inorganics?
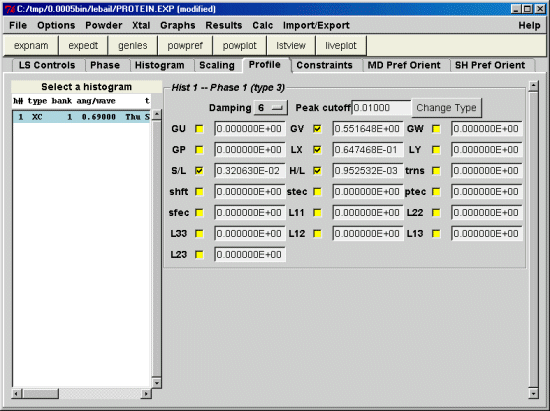
Yes, one needs to go to the least squares controls menu &
pick a band width for the LS matrix; I use 300 for my work
& I know Jon Wright has used 50. Also pick a Marquardt
damping factor; try 1.20 to start & adjust as needed. Try
to make small if possible. Finally, change the convergence
factor; you enter this as the log, so -2.0 is the default
of 0.01. For a 3000 parameter protein refinement 1.7 (~50
for sum shift/esd) is more reasonable. I also set the
print option for shift/esd summary table and do 9 cycles
at a time.
>I take it after the structure is happily imported, it is
>just a matter of running all the restraints macros in
>the c:\gsas\macros directory (except C60)
Yes, these will go through the protein structure & build
all the needed restraints. This is pretty quick. All the
amino acids in the protein must be one of the standard 20.
You'll have to do other molecules (ligands, etc.) by hand.
A few other pointers: I use Swiss PDB Viewer to look at
the resulting crystal structure. It reads PDB files -
these are made by gsas2pdb. SPDBV can then be used to fix
up "bad" bits in the structure - "mutate" side chains,
etc., save the result and GSAS can then read back in the
new coordinates. One careful point is to change all the
UISO's back to 0.3 (or whatever you pick) as SPDBV tends
to change them to nonsense if the "mutate" is used.
To: [email protected]
From: Jonathan WRIGHT [[email protected]]
Subject: Re: RIET: GSAS query on importing Protein PDB atom positions?
> >Would you normally also import the water/HOH molecules,
> >disordered
> >atoms and hydrogens?
> >(though the PDB file I seem to have does not include the
> >hydrogens)
>
>"Normally" not for powder data. Single crystal protein
>folks seem only to add waters with higher resolution data
>(dmin ~ 2.5A or better). Some PDB files do have H-atoms. I
>change the UISO's for all the atoms to 0.3 which is about
>average for protein atoms. Powder data isn't good enough
>to do UISO's in a refinement even when all constrained to
>be identical.
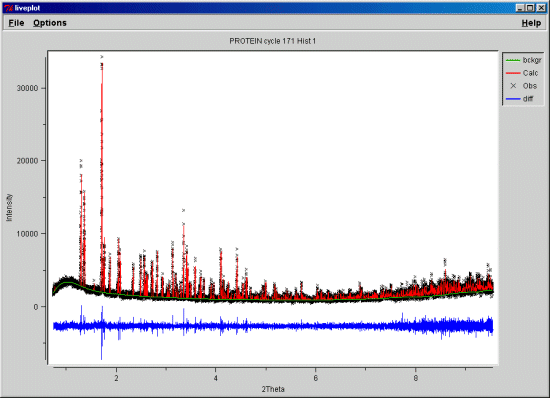
In the case which I think Lachlan is looking at - you have a ~1.5 angstrom
(=very good) single crystal structure, which matches the powder data.
Importing or not the water molecules should only really impact on the
solvent scattering factors, but if you want to refine the structure it will
be something of a challenge to get the waters to keep to sensible
positions. If you delete the the H2O/H parts of the structure then in
theory you'll just put them back into the structure via the "solvent
scattering", but not know exactly where they are any more. For neutron data
(if you have any), it ought to make a much bigger difference, as the
simplistic solvent scattering model has been reported to break down with
single crystal data. Remember that refining a high resolution single
crystal structure against powder data is only going to make it worse,
whether it's a protein, inorganic structure or small organic molecule,
assuming the sample is the same.
A question for Bob, have you (or anyone else) tried fitting any single
crystal datasets using GSAS? In theory it ought to be an alternative to
shelx, refmac etc?
Cheers,
Jon
From: "Bob Von Dreele" [[email protected]]
Subject: Re: RIET: GSAS query on importing Protein PDB atom positions?
To: [email protected]
Date: Tue, 03 Dec 2002 08:28:20 -0800
On Tue, 03 Dec 2002 13:54:53 +0100
Jonathan WRIGHT [[email protected]] wrote:
>
>>>Would you normally also import the water/HOH molecules,
>>>disordered
>>>atoms and hydrogens?
>>>(though the PDB file I seem to have does not include the
>>>hydrogens)
>>
>>"Normally" not for powder data. Single crystal protein
>>folks seem only to add waters with higher resolution data
>>(dmin ~ 2.5A or better). Some PDB files do have H-atoms.
>>I change the UISO's for all the atoms to 0.3 which is about
>>average for protein atoms. Powder data isn't good enough
>>to do UISO's in a refinement even when all constrained to
>>be identical.
>
>In the case which I think Lachlan is looking at - you
>have a ~1.5 angstrom (=very good) single crystal
>structure, which matches the powder data. Importing or
>not the water molecules should only really impact on the
>solvent scattering factors, but if you want to refine the
>structure it will be something of a challenge to get the
>waters to keep to sensible positions. If you delete the
>the H2O/H parts of the structure then in theory you'll
>just put them back into the structure via the "solvent
>scattering", but not know exactly where they are any
>more. For neutron data (if you have any), it ought to
>make a much bigger difference, as the simplistic solvent
>scattering model has been reported to break down with
>single crystal data. Remember that refining a high
>resolution single crystal structure against powder data
>is only going to make it worse, whether it's a protein,
>inorganic structure or small organic molecule, assuming
>the sample is the same.
This is an interesting problem (single crystal vs powder).
I've not found that a calculated powder pattern from a
protein single crystal structure can actually reproduce
the observed powder diffraction pattern. The discrepancies
are most apparent in the low angle part (>8A d-spacing)
and are quite large. Because the size of the d-spacings
(10-30A) for these reflections and the magnitude of the
differences, they are associated with very large scale
features in the crystal structure and not just the
placement of a hundred or so water molecules.
Consequently, Rietveld refinement of the protein structure
starting with a "good" single crystal structure will show
shifts in atom positions of 1-2A. This also means that
trying to do joint single crystal-powder refinements don't
seem to work very well. I've often wondered if the powder
material (1 micron crystals) may in fact be inherently
different from the usual single crystals used for protein
work (100+ micron crystals). I can't tell from comparisons
of lattice parameters because the precision of the single
crystal values isn't really good enough to make any useful
comparison to the powder ones. Any comments?
>A question for Bob, have you (or anyone else) tried
>fitting any single crystal datasets using GSAS? In theory
>it ought to be an alternative to shelx, refmac etc?
I have. It works just fine. The PDB (www.rcsb.org) does
have structure factors deposited for many of the protein
structures. These can be read into GSAS in the usual way
and the protein structure refined with as many restraints
as one wishes. However, GSAS doesn't "do" R(free), etc.
familiar to the protein folks. Results aren't any
different that those obtained by conventional protein
programs (TNT, O, etc.).
Bob
Date: Tue, 03 Dec 2002 18:38:51 +0100
To: [email protected]
From: Jonathan WRIGHT [[email protected]]
Subject: Re: RIET: GSAS query on importing Protein PDB atom positions?
>This is an interesting problem (single crystal vs powder).
>I've not found that a calculated powder pattern from a
>protein single crystal structure can actually reproduce
>the observed powder diffraction pattern. The discrepancies
>are most apparent in the low angle part (>8A d-spacing)
>and are quite large.... Any comments?
For the myoglobin I found that the deposited observed structure factors
match the powder pattern fairly well (but not exactly). Most of the
problems were due to the dataset being incomplete below about 10 angstrom,
so any comparison for those peaks was not possible. I seem to remember that
the deposited data did a better job than the deposited structure for the
low resolution data. There is some neutron single crystal work on myoglobin
which developed a more sophisticated model for the solvent scattering than
the Babinet's principle model (x-ray models were really hopeless for
neutron data at low resolution). I have the impression that being lousy at
low resolution is a known problem with many x-ray models of protein
structure, which various people have improved by modelling the solvent in
more sophisticated ways. Unfortunately it's the intensities which are
easiest to resolve with a powder which are the hardest to account for!
Cheers,
Jon
From: "Bob Von Dreele" [[email protected]]
Subject: Re: RIET: GSAS query on importing Protein PDB atom positions?
To: [email protected]
Date: Tue, 03 Dec 2002 10:43:02 -0800
On Tue, 03 Dec 2002 18:38:51 +0100
Jonathan WRIGHT [[email protected]] wrote:
>>This is an interesting problem (single crystal vs
>>powder).
>>I've not found that a calculated powder pattern from a
>>protein single crystal structure can actually reproduce
>>the observed powder diffraction pattern. The
>>discrepancies
>>are most apparent in the low angle part (>8A d-spacing)
>>and are quite large.... Any comments?
>
>For the myoglobin I found that the deposited observed
>structure factors match the powder pattern fairly well
>(but not exactly). Most of the problems were due to the
>dataset being incomplete below about 10 angstrom, so any
>comparison for those peaks was not possible. I seem to
>remember that the deposited data did a better job than
>the deposited structure for the low resolution data.
>There is some neutron single crystal work on myoglobin
>which developed a more sophisticated model for the
>solvent scattering than the Babinet's principle model
>(x-ray models were really hopeless for neutron data at
>low resolution). I have the impression that being lousy
>at low resolution is a known problem with many x-ray
>models of protein structure, which various people have
>improved by modelling the solvent in more sophisticated
>ways. Unfortunately it's the intensities which are
>easiest to resolve with a powder which are the hardest to
>account for!
Jon,
It's also my understanding that these reflections are also
the most difficult to measure accurately by the single
crystal techniques used in protein data collection. I also
wonder if the hydration models used for protein structure
analysis take on the same role as anisotropic thermal
parameters do in small molecule work (i.e. as a "catch
all" for systematic errors!). I also see that the
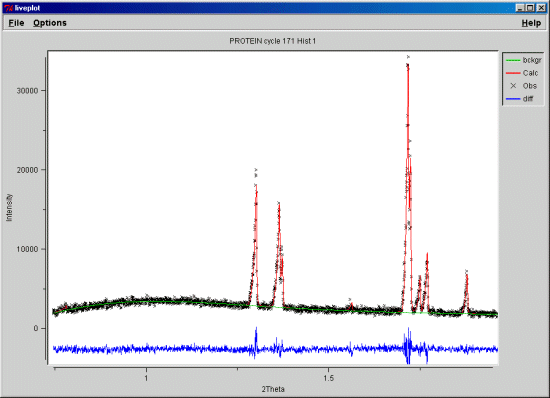
calculated powder pattern from a single crystal protein
structure does match the powder pattern for higher 2-theta
(d<6A).
Bob