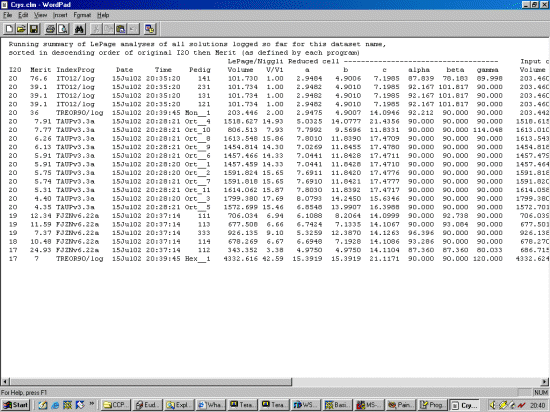
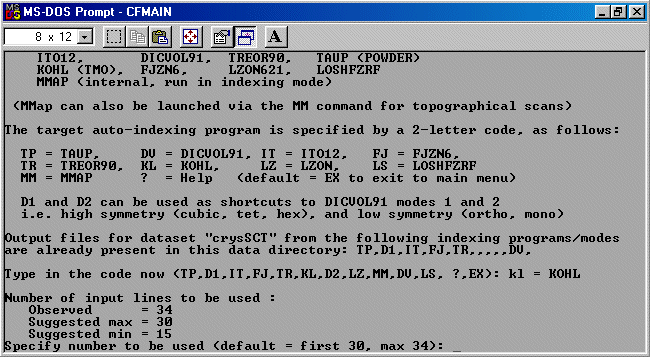
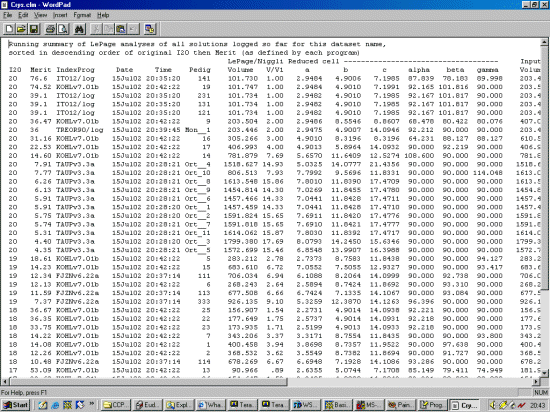
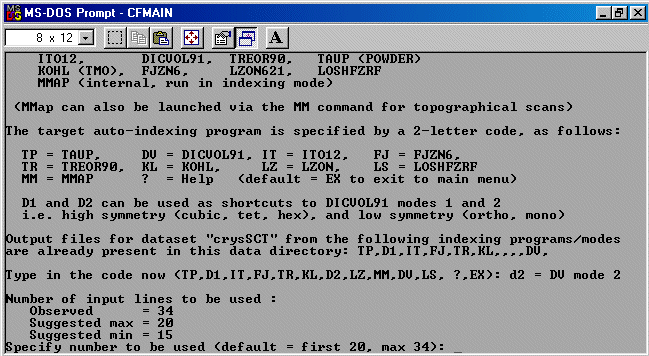
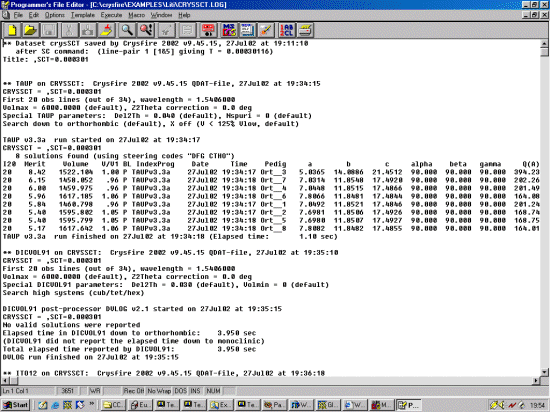
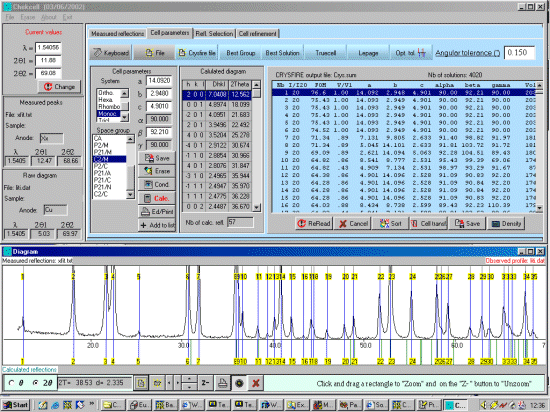
Note: The following is just a single example of using CRYSFIRE. There are other
methods of interacting with the program and doing the following. The assumption is that
you have got your peaks / observations into a Crysfire CDT file (via some of the other tutorials).
This run through will use the peak list file obtained in the
Lithium Titanate Fundamental
Parameters Peak Profiling Tutorial. (I.E. Grey, L. M. D. Cranswick, C. Li,
L. A. Bursill, and J. L. Peng, "New Phases Formed in the Li-Ti-O System under
Reducing Conditions", Journal of Solid State Chemistry, 138, 74-86 (1998))
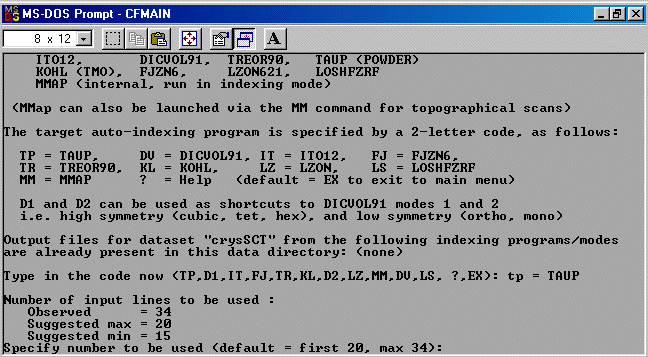
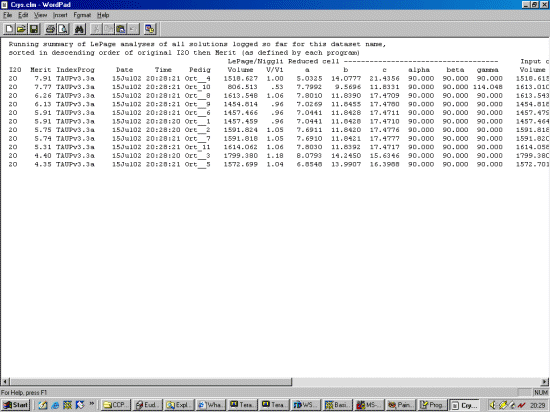
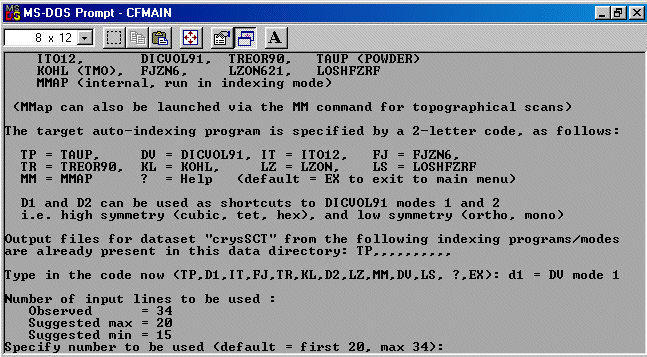
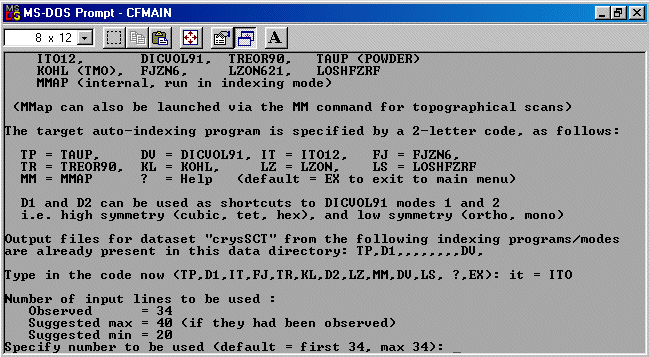
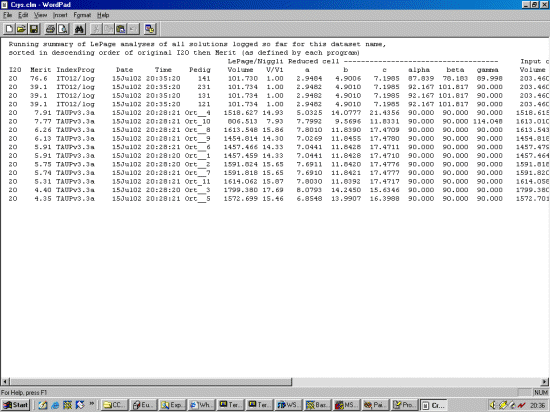
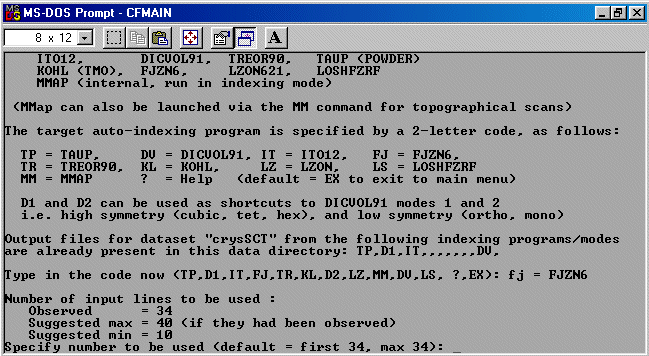
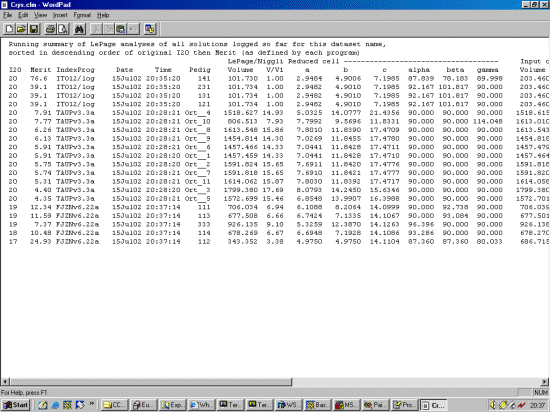
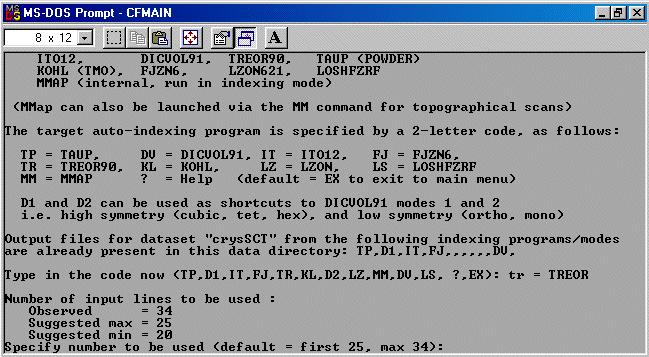
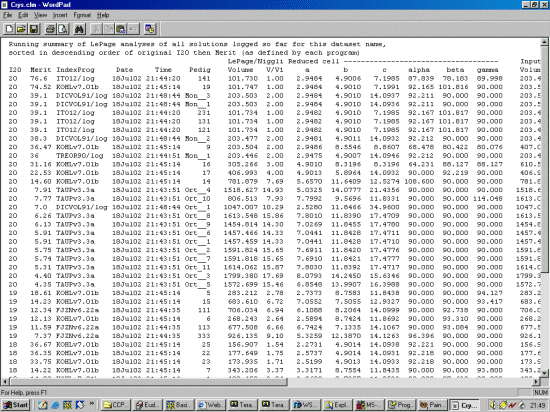
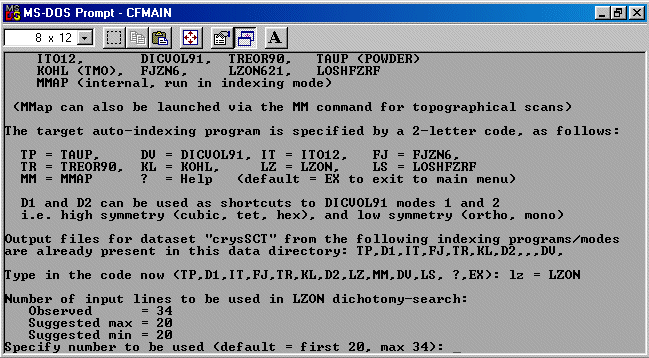
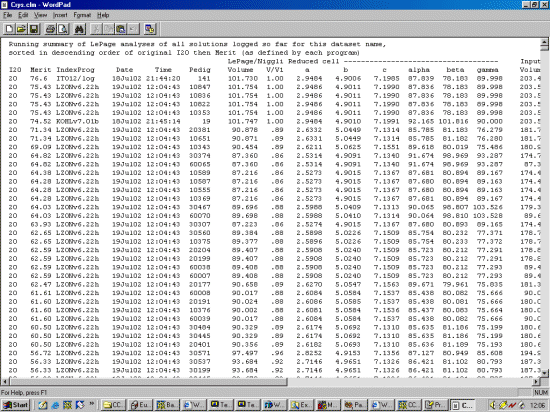
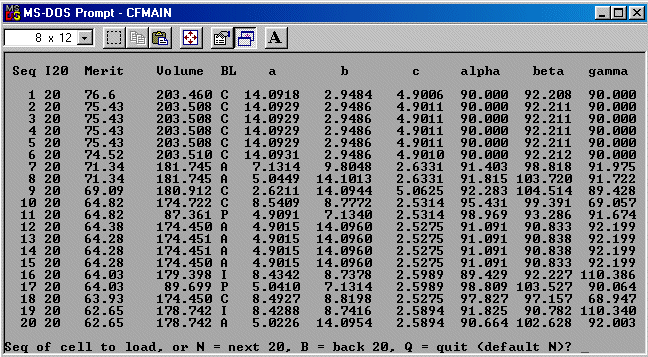
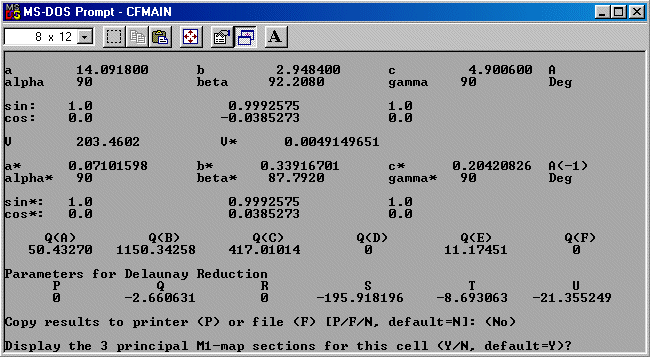
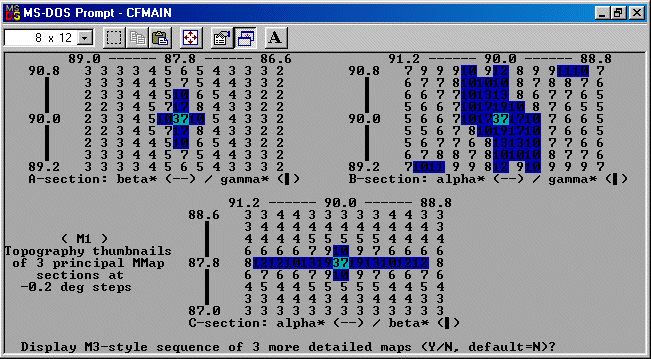
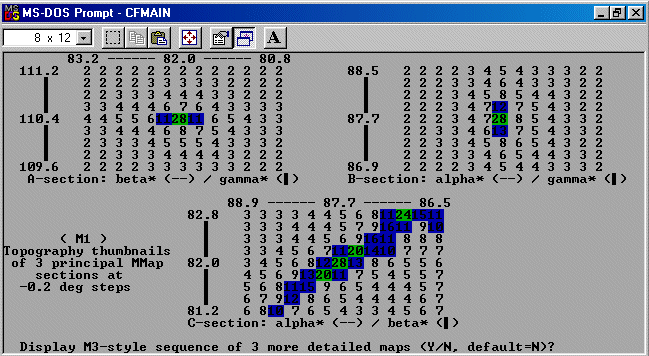
The strategy here is INdex using all the available programs by using
the IN command. By using all the programs, we will then obtain a good idea
at the range of possible solutions the data will support. Hopefully one or two
of those solutions will appear superior compared to the great mass of other trial cells.
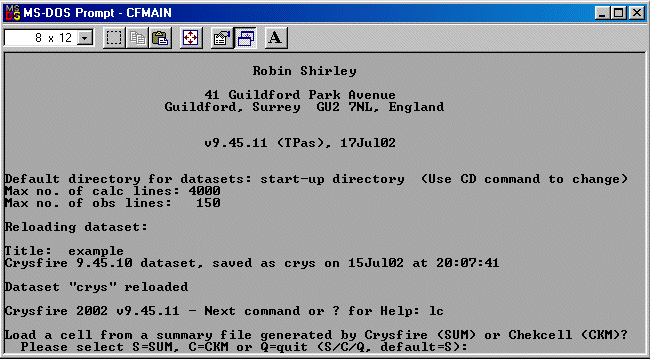
The following assumes you have already created a Crysfire CDT file. Example tutorials for
importing data into Crysfire are:
A comment from Robin Shirley about filenames and descriptive text
From: Robin Shirley [[email protected]]
Organization: Psychology Dept, Surrey Univ. U.K.
To: Lachlan Cranswick [[email protected]]
Date: Mon, 22 Jul 2002 18:43:55 GMT
Incidentally, in this context, could you please emphasise in your
tutorials the importance of spending the few seconds that it takes to
give each new dataset a well-chosen name and some helpful brief
descriptive text in its description field, seeing that whatever is used
there will remain the default throughout the rest of the analysis?
Careful dataset naming really is important for keeping track of progress
in any serious study that can involve several dataset variants, and
such distinctive names have been assumed throughout Crysfire as the basis
of its data organisation.
Users shouldn't rely just on having kept each problem separate within
its own data directory (folder).
While that's important, it's *not* sufficient, because (a) doing that
only identifies the directory, not the files within it, and (b) as the
study progresses, it's likely that several datasets and dataset variants
will be required, for example after recalibration, applying estimated
Z2theta or specimen-displacement corrections, rescaling, etc.
These will all need to be kept in the same directory so that, for
example, their trial cells can easily be loaded and examined (LC, M1,
etc), but if they aren't given different names (and preferably also
different description fields - see below) then complete confusion will
quickly result.
Since the description field gets appended to every summary-file solution
line, an important opportunity is lost if its contents can't act as an
aide memoire of the characteristics of the particular dataset or dataset
variant that was used for that trial solution.
This becomes particularly relevant when datasets are merged and/or
rescaling/unscaling is used, as was discussed above and as is
increasingly likely to happen in response to the new volume estimates
and rescaling prompts in CF2002.
I'm already regretting that I short-sightedly left it far too easy in
WF2crys and XF2crys for users to default to an undistinctive and
meaningless dataset name like Crys.cdt (I've noticed that you are
yourself a frequent offender in this regard!), and I may well look at
ways to prevent this in the next revisions of those programs.
As things stand, it's too easy for a moment's impatience on the part of
the user at this basic choice point to handicap all subsequent work on
that sample with a basically null name (and description) - I really
should have had the foresight to protect users from this pitfall.
(A bit of a rant, perhaps, but I do think the point needs making.)
With best wishes
Robin