A program for preparing VRML-1 files for 3D visualization of RMCA modelling
by Armel Le Bail (April - 1997)
Version 1.0 (32 bits) for PC
under Windows 95 and probably NT
DOWNLOAD GLASSVIR.ZIP
(updated April 18, 1997)
Content
Purpose of the program
Running the program
Result as seen by VRweb
Warnings
To do next
To read a configuration file (.cfg) from a RMCA modelling (glass or other), then to create 3D VRML-1 files (.wrl) containing polyhedra for a selected atom pair, which can be examined further with a VRML viewer like VRweb or Live3D (see below). This program is better suited for materials with exclusive environments (oxides, fluorides ...).
For information on RMCA, see : http://www.studsvik.uu.se/rmc/rmchome.htm
Some Window 95 executables are available (RMCA,
RANDOM, MOVEOUT, CRYSTAL and NOCHAOS)
For information on VRML as related to crystallography see : http://www.cristal.org/vrml/intvrml.html
Recommended viewers for the VRML-1 files are :
VRweb : a standalone viewer available at http://www.iicm.edu/vrweb
Live3D : the plug-in delivered with Nestcape 3 or 4
The glassvir.zip file contains
The program is executed either by a double click on glassvir.exe from the explorer or inside a DOS box opened in the directory containing it by typing "glassvir".
The configuration file (.cfg) to be examined should be in the same directory as the executable.
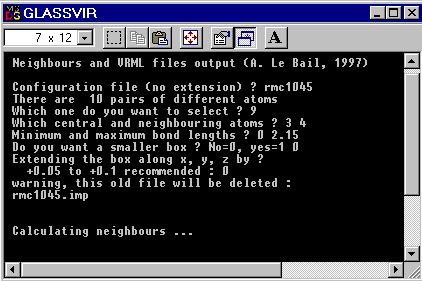
When the program is started, you will have to give responses to a few questions, like for the following rmc1045.cfg test file :

If you had choosen a smaller box, the dialog would be :

Then the DOS box is closed automatically at the end of calculations (if it was opened by a double click from the Windows Explorator).
Files (all ASCII) will be created in the same directory :
Content of the file rmc1045.imp :
-------------------------------------------------------------------------- Calculation of neighbours in rmc1045
No. of atom types = 4
Minimum bond lengths = 0.00000000E+000 Maximum bond lengths = 2.15000010
Type 3 - Type 4 neighbours:
6 300 100.000
Average coordination 6.00 --------------------------- --------------------------------------------------------------------------
The test file corresponds to a fluoride glass with formulation NaPbFe2F9, with 150 Na and Pb atoms, 300 Fe atoms and 1350 F atoms, in this order in the rmc1045.cfg configuration file :
Atom type : 1 2 3 4
Na Pb Fe F
Thus there are 10 atom pairs which are classified as :
Atom pairs : 1 2 3 4 5 6 7 8 9 10
Na-Na Na-Pb Na-Fe Na-F Pb-Pb Pb-Fe Pb-F Fe-Fe Fe-F F-F
So that if you want to observe the Fe coordination with fluorine atoms, you should select the ninth atom pair and this corresponds to the central and neighbouring atoms number 3 and 4. This should clarify the responses given to the questions asked above and recalled below :
which one do you want to select ? 9 which central and neighbouring atoms ? 3 4 Minimum and maximum bond lengths ? 0 2.15
These bond lenghts allow to find exclusively FeF6 polyhedra in this particular case.
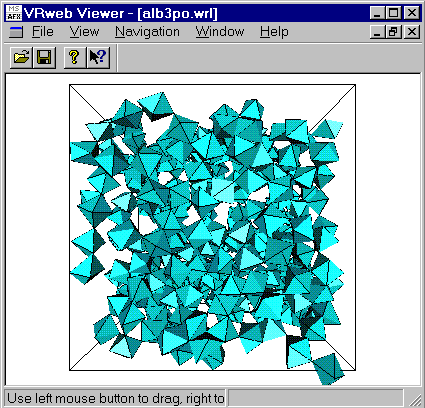
If you have yet a VRML viewer, load the po.wrl file renamed
as alb3po.wrl
(CAUTION, it is
big (208K) with 300 octahedra in a cubic box having 30.12 Angstroms length)
More on the NaPbFe2F9 case at : http://www.cristal.org/glasses/napbm2f9.html
GLASSVIR was built on the basis of STRUVIR for which a tutorial and lot of informations are available at : http://www.cristal.org/vrml/struvir.html
Armel Le Bail
Laboratoire des Fluorures, CNRS UPRES-A 6010
Universite du Maine - Faculte des Sciences
Avenue Olivier Messiaen
72085 LE MANS Cedex 9 FRANCE
E-Mail [email protected]
Web http://www.cristal.org/