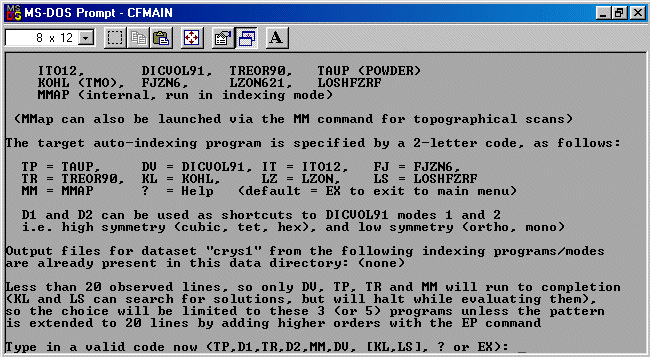
Performing Pattern Extension in Crysfire
To extend the pattern to 20 peaks, run the EP (Extend Pattern) command.
Crysfire recommends and prompts the saving of the modified data to a CDT file with EP
to the filename (e.g., in this case, crys1ep.CDT).
CDT file BEFORE pattern extension
"CRYS v9.30c dataset, saved as crys1 by XF2CRYS v1.7b on 27Jul02 at 22:04:33"
""
"To use with CRYS versions prior to v9.30, delete these first 3 lines"
"crys1",7,0,0,0,0,1.54056,0,1,"P"
0,0,0,0,28.3464,4837.0742
0,0,0,0,47.1625,1766.6555
0,0,0,0,55.976,1277.9629
0,0,0,0,68.95010000000001,297.0331
0,0,0,0,76.22929999999999,627.768
0,0,0,0,87.8663,497.7363
0,0,0,0,94.7882,0
CDT file AFTER pattern extension
"Crysfire 9.45.15 dataset, saved as crys1EP on 27Jul02 at 22:19:52"
",EP20"
"To use with CRYS versions prior to v9.30, delete these first 3 lines"
"crys1EP",20,0,0,0.0000000000000,0,1.00000000,0.0000000000000,0,"P"
1010.4546292934787, 0, 0, 0, 18.2905470936284,4837.07400000
2697.3010526759795, 0, 0, 0, 30.1018819434549,1766.65500000
3711.7534113792990, 0, 0, 0, 35.4707042567356,1277.96200000
5400.1797717844256, 0, 0, 0, 43.1143594758309,297.03300000
6421.0603898682257, 0, 0, 0, 47.2385889019994,627.76000000
8113.2459397443172, 0, 0, 0, 53.5345671495925,497.73600000
9130.4188302460898, 0, 0, 0, 57.0792110165363, 0
4041.8185171739146, 0, 0, 0, 37.0691050114265, 0
9094.0916636413076, 0, 0, 0, 56.9551462930426, 0
10789.2042107039178, 0, 0, 0, 62.5780553697409, 0
14847.0136455171960, 0, 0, 0, 75.0693734135128, 0
16167.2740686956586, 0, 0, 0, 78.9517079662539, 0
21600.7190871377024, 0, 0, 0, 94.5906323740015, 0
24275.7094740838147, 0, 0, 0,102.3442834511437, 0
25261.3657323369662, 0, 0, 0,105.2522015900592, 0
25684.2415594729027, 0, 0, 0,106.5117564386302, 0
32452.9837589772687, 0, 0, 0,128.5098513240440, 0
33405.7807024136928, 0, 0, 0,132.0893765605785, 0
36376.3666545652304, 0, 0, 0,144.9666786397511, 0
36521.6753209843591, 0, 0, 0,145.6985228337022, 0